Are you making this mistake? How to perform data normalization in your models.
If you have been working with Scikit-learn for a while you probably already heard somewhere about how much better and safer a pipeline is. Pipeline is a Scikit-learn function used to aggregate all the steps between data pre-processing and model training/optimization.
In this post I will reference the functions and libraries in the figure below, you can find the whole code HERE:

One of the main advantages of using a pipeline is for data standardization. Data standardization is an important step for many machine learning classifiers. For some distance-based ones, like the Support Vector Machine (SVM), it is of utmost importance to make sure that the computed distances between features are not dominated by large values. For others, like Neural Networks, can severely improve training and converging time.
It is good practice to split the data into a train and test set prior to using a standardization method. The method is trained using the training set and with the transform function, is applied to the test set. This prevents data leakage from the training set into the testing set, which can severely bias our results. See the figure below for the code example.

Often, the next step is to find the best parameters for our classifier. For that, we often use a form of grid search, either the RandomizedSearchCV or GridSearchCV. See the figure below for an example.

Now, here’s a problem that not using a pipeline will raise. GridSearch will use only the training set and will split it into N folds (N is given by the cv parameter). N-1 folds are user to train and the remaining 1 fold to test and see how good a given combination of hyper-parameters is. But, when we standardized the data, we did not take into account the splits (inner cross-validation) that would happen in the GridSearch step. We used our whole training set to train the standardizer, therefore, the splits generated during GridSearch are not 100% independent. The model is optimizing hyper-parameters on a train and validation split where data leakage was introduced, and therefore, our optimization results might be biased.
Your test set is not really affected by this small problem, but the hyper-parameters chosen during GridSearch might not generalize as well in the testing set, since the process to find them was not 100% trustworthy.
Now, this could be easily avoided if we used the Pipeline class from Scikit-learn. In the Figure below, we define each of the steps that will happen in the pipeline during GridSearch (Standardization and the classifier to be optimized).
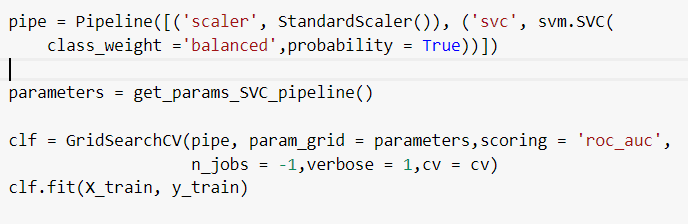
The approach above will guarantee that the splits inside GridSearch are done in a proper way, preventing data leakage.
Now, what is the actual effect of using or not a pipeline? To show this, I created 2 experiment settings using the Pima Indians Diabetes Database (from a Kaggle challenge: available here) and the Breast Cancer Wisconsin (Diagnostic) Data Set (also from Kaggle: available here).
For the diabetes dataset, the objective is to predict whether a given patient has diabetes or not. For the breast cancer dataset, the objective is to classify cancer into malignant or benign.
I split both data sets using 5 fold cross-validation and for each k-fold iteration, I standardized the data with and without a pipeline and optimized the hyper-parameters. For each k-fold iteration, I recorded the area under the receiver operating characteristic curve (AUC)for the test set and the hyper-parameters resulted from the grid-search.
Here’s the code so you can follow it easily (to run it in Colab you will need to download the data from Kaggle in the links provided above):
Results — Pima Indians Diabetes Database


Results — Breast Cancer Wisconsin


As you can see, I could not find any differences between using a pipeline or normalizing the data separately. The AUC and the hyper-parameters chosen were exactly the same for both datasets. That could be, of course, due to the datasets themselves (which are not very complex) or to the chosen model *.
That being said, the theoretical reasoning behind the use of the pipeline function is unquestionably correct and its use will make your code not only cleaner and easier to maintain, but also safer against over-optimistic results.
*Do you think there’s something wrong in the code? Is there another experiment you would like to see? Leave a comment or send me an e-mail. l.a.ramos@amsterdamumc.nl
